Introduction
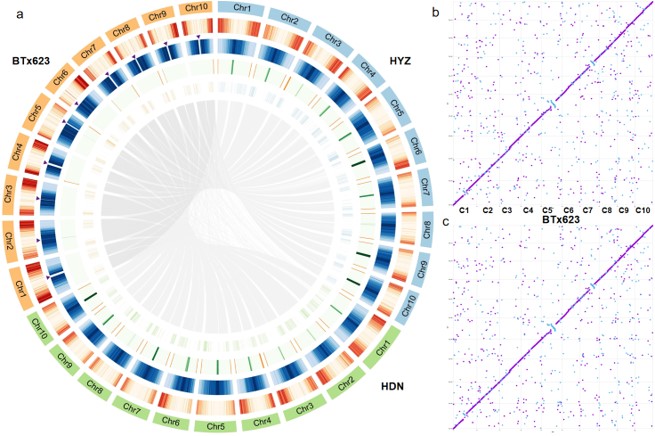
The use of sorghum to brew Baijiu, a traditional distilled liquor, has a history of thousands of years in China. These Baijiu-brewing sorghums are waxy and tannin-rich, exhibit distinct phynotypes from the reference genome inbred BTx623. However, little is known about the genome of these landraces. Here we present the complete telomere-to-telomere (T2T) genome assemblies of the representative Chinese Baijiu-brewing sorghum cultivar Hongyingzi (红缨子 for Maotai, the most famous Baijiu in China) from Chishui River basin and
Huandiaonuo (环雕糯, for Fen-flavor Baijiu) from Yangtze River basin, integrating PacBio HiFi, 100-kb ultra-long ONT (UL-ONT), and Hi-C sequencing technologies.

Summary of T2T genome assembly of Chinese Baijiu-brewing sorghum
BTx623
Hongyingzi
Huandiaonuo
Draft assembly size (Mb)
708.74
745.65
738.5
Contigs
4773
342
241
Contig L50
78
5
5
Contig N50 (Mb)
1.3
70.87
66.59
Chromosome Genome size (Mb)
683.65
724.37
726.89
Chromosomes
10
10
10
Gaps
2679
0
0
Percaentage Repeat sequence
64.89 %
70.11 %
70.04 %
Telomeric repeats (CCCTAAA)n
14/4 paired
20/10 paired
20/10 paired
BUSCO
98.10%
99.50%
98.63%
Base quality value score(QV)
>50
70.11
66.76
Protein-coding genes
34,118
43,912
44,465